Unusual nucleic acid structures
More than 60 years after the discovery of the double helix, we have yet to grasp the structural complexity of nucleic acids. These biomolecules cannot be viewed as simple single- or double-stranded linear polymers, but rather as complex 3D architectures. We are interested in two non-canonical conformations of four-stranded nucleic acids, the i-motif and the guanine quadruplexes (G4):
The i motif, discovered in 1993 at Ecole Polytechnique, is an unusual architecture, based on semi-protonated C.C+ base pairs. This structure then fell into relative oblivion due to its instability at physiological pH, but we recently showed that a few sequences could be folded in the cell thanks to in cell NMR. Our recent work has made it possible to measure and predict their stability in vitro and to analyze their spectroscopic properties.
The guanine quadruplexes are peculiar three-dimensional structural arrangements adopted by G-rich DNA and RNA strands. They result from the stacking of several guanine quartets (linked by hydrogen bonds between 4 coplanar guanines) and stabilized by cations such as K+, which are very abundant in the cell. Cations also play an essential role in the structure and stability of G4s. (Figures 1-2).
Fig. 1. i-DNA (left) vs G-quadruplexes (right). (A) i-DNA is exemplified here with PDB 1YBL. (B) The hemiprotonated C·C+ base pair. The N3 atom of one cytosine must be protonated for the formation of a third H-bond (red). (C,D) A G-quadruplex is based on the formation of G-quartets.
Research into quadruplexes has sustained its momentum because, contrary to many other alternative DNA or RNA conformations, G4 structures are stable under physiological conditions. The driving forces for duplex opening may be related to replication and transcription events, molecular crowding, and stabilization of transient single-stranded regions by proteins. G4-prone sequences are distributed throughout eukaryotic genome in promoters, immunoglobulin switch regions, ribosomal and mitochondrial DNA. Interestingly, G4 motifs can also be found in prokaryotes and viruses. Critical evidence has also emerged for a variety of prokaryotes (e.g. bacteria such as Neisseria gonorrhoeae or Archaea such as Haloferax volcanii) or eukaryotes, explaining why G4 have gone from being a mere laboratory curiosity to being a "hot topic".
There is little doubt that this structure is formed at least transiently in vivo and constitutes a challenge for the replication, transcription or repair machineries, but also a regulatory element modulating key events.
We are developing biophysical methods to study these structures in vitro. In addition, as the available prediction tools appearing insufficient to us, we have undertaken to develop ourselves a novel, more reliable prediction algorithm, G4-Hunter, which facilitates genomic analyzes which is now used worldwide.
Cancer: We are studying the involvement of G-quadruplexes in cancer-related processes. We and others have initially developed G4-ligands as telomerase inhibitors or agents interfering with telomere capping. These ligands have an antiproliferative effect on cancer cells. Nevertheless, we showed that the TRAP assay is not suited, at least in its classical format, to investigate G4 ligands: these compounds cannot be considered as "simple" telomerase inhibitors, leading to a paradigm shift in the understanding of the functions of these molecules. Besides the extremities of eukaryotic chromosomes, G4-prone motifs are often found in the promoters of human genes, and especially abundant in the promoters of oncogenes such as KRAS or B-MYB.
Viruses: We are investigating the antiviral effects of G4 ligands: many viruses exhibit conserved G4 patterns that can be targeted. Our analysis of pool of 11,000 accessible viral genomes revealed that viruses causing acute infections are significantly depleted for quadruplex motifs. We also recently showed that the SUD domain of the Nsp3 protein of the SARS-CoV2 virus recognizes RNA quadruplexes, and that it was possible to inhibit this interaction with small ligands, which have antiviral activity on cells in culture. (https://www.polytechnique.edu/actualites/vers-une-nouvelle-strategie-antivirale-contre-la-covid-19).
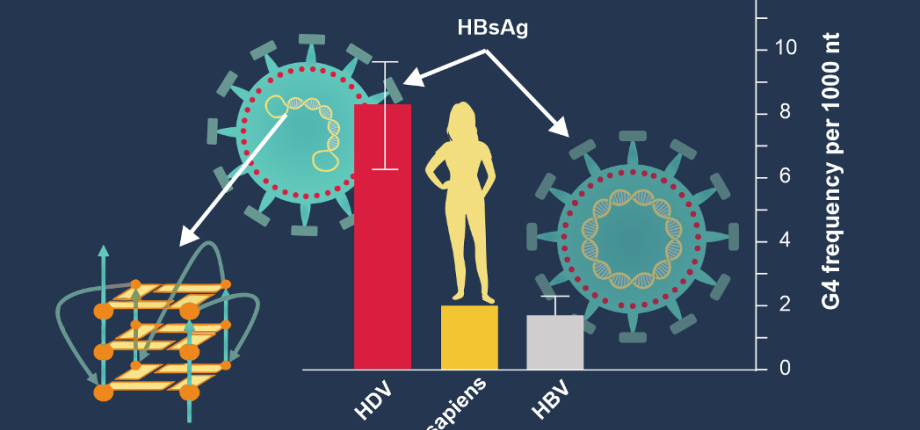
Paleogenomics: We have shown shown that the hepatitis B virus (HBV) has increased the frequency of G4s in its genome over the past 10,000 years to better blend with the human genome and continue to benefit from its cellular machinery to multiply and survive (https://www.ip-paris.fr/actualites/des-structures-particulieres-de-ladn-eclairent-levolution-du-virus-de-lhepatite-b). This paleogenomic work – the study of ancient genomes – is now being continued by the analysis of the genome of Neanderthals, our “cousin” who disappeared around 42,000 years ago... Understanding the evolution of our genomes also tell us about the history of the fight against certain pathogens: their genome evolves, but so do our defenses!
Parasites: Besides viruses, we are interested in parasites such as Plasmodium falciparum, responsible for malaria, Trypanosoma cruzi, responsible for Chagas disease, certain nematodes or finally a platyhelminth parasite of humans, Schistosoma mansoni (https://www.polytechnique.edu/actualites/cibler-des-structures-inhabituelles-de-ladn-contre-les-maladies-parasitaires)
Our expertise on G4s is now recognized worldwide, in particular in the development of methods for analyzing their stability and structure, We welcome many visitors from all over the world who to come to test their sequences or ligands. Conversely, we have established a strong network of national and international collaborations and actively participated in the creation of a learned society, the G4 society (https://www.g4-society.org). It is interesting to underline that the French community in this field is very active, both in chemistry and in biology, and that it was structured thanks to annual scientific meetings and networks (GDR, COST) to which we contributed.
Fig. 2. Selected front cover of past publications
----------------------------------------------------------------------------------------
Selected Recent publications:
Gajarsky M, Stadlbauer P, Sponer J, Cucchiarini A, Dobrovolna M, Brazda V, Mergny JL, Trantirek L, and Lenarcic Zivkovic M. DNA quadruplex structure with a unique cation dependency Angew Chem Int Ed Engl. (2024) 12, e202313226.. https://doi.org/10.1002/anie.202313226
Víšková P, Ištvánková E, Ryneš J, Džatko S, Loja T, Lenarčič Živković M, Rigo R, El-Khoury R, Serrano-Chacón I, Damha MJ, González C, Mergny JL, Foldynová-Trantírková S, Trantírek L. In-cell NMR suggests that DNA i-motif levels are strongly depleted in living human cells Nature Comm. 2024, 15, 1992. https://doi.org/10.1038/s41467-024-46221-y
Luo Y, Lenarčič Živković M, Wang J, Jan Ryneš J, Foldynová-Trantírková S, Trantírek L, Verga D, & Mergny JL. A sodium-potassium switch for G4 prone G/C-rich sequences. Nucleic Acids Res. (2024) 52, 448-461. https://doi.org/10.1093/nar/gkad1073
Brázda V, Valkovia N, Dobrovolná M, Mergny JL. Abundance of G-quadruplex forming sequences in the Hepatitis Delta virus genomes. ACS Omega (2024) 9, 4096-4101. https://doi.org/10.1021/acsomega.3c09288
Esnault C, El Aabidine AZ, Garcia-Oliver E, Cucchiarini A, Lleres D, Goerke L, Luo Y, Verga D, Lacroix L, Feil R, Spicuglia S, Mergny JL, Andrau JC. G4access reveals G-quadruplexes association to open chromatin and to imprinting regions control. Nature Genetics (2023) 55, 1359-1369. https://doi.org/10.1038/s41588-023-01437-4
Skolakova P, Gajarsky M, Palacky J, Šubert D, Renčiuk D, Trantirek L, Mergny JL, Vorlíčková M. DNA i-motif formation at neutral pH is driven by kinetic partitioning. Nucleic Acids Res. (2023) 51, 2950-2962. https://doi.org/10.1093/nar/gkad119
Brázda V, Dobrovolná M, Bohálová N, Mergny JL. G-quadruplexes in the evolution of hepatitis B virus. Nucleic Acids Res. (2023) 51, 7198–7204. https://doi.org/10.1093/nar/gkad556. Commented by CNRS and IP Paris: https://www.ip-paris.fr/actualites/des-structures-particulieres-de-ladn-eclairent-levolution-du-virus-de-lhepatite-b
Zhang X, Qiu D, Chen J, Zhang Y, Wang J, Chen D, Liu Y, Cheng M, Monchaud D, Mergny JL, Ju H, Zhou J. A Chimeric Biocatalyst Combining Peptidic and Nucleic Acid Components Overcomes the Performance and Limitations of the Native Horseradish Peroxidase. J Am Chem Soc. (2023) 145, 4517-4526. https://pubs.acs.org/doi/10.1021/jacs.2c11318
Zhang R, Shu H, Wang Y, Tao T, Tu J, Wang C, Mergny JL*, Sun X. G-quadruplex structures are key modulators of somatic structural variants in cancers. Cancer Research. (2023) 83, 1234-48. https://doi.org/10.1158/0008-5472.CAN-22-3089
Ferret L, Alvarez-Valadez K, Rivière J, Muller A, Bohálová N, Luo Y, Guittat L, Brázda V, Kroemer G, Mergny JL*, Djavaheri-Mergny M. G-quadruplex ligands as potent lysosome regulators. Autophagy (2023) 19, 1901-1915. https://doi.org/10.1080/15548627.2023.2170071
Vinayagamurthy S, Bagri S, Mergny JL, Chowdhury S. Telomeres Expand Sphere of Influence: Emerging Molecular Impact of Telomeres in Non-telomeric Functions. Trends in Genetics (2023) 39, 59-73. https://doi.org/10.1016/j.tig.2022.10.002
Figueiredo J, Djavaheri-Mergny M, Ferret L, Mergny JL, Carla Cruz. Harnessing G-quadruplex ligands for lung cancer treatment: a comprehensive overview. Drug Discovery Today (2023) 28, 1-18. https://doi.org/10.1016/j.drudis.2023.103808
Luo Y, Granzhan A, Marquevielle J, Cucchiarini A, Lacroix L, Amrane S, Verga D, Mergny JL. Guidelines for quadruplexes. I. in vitro characterization Biochimie (2023) 214, 5-23. https://doi.org/10.1016/j.biochi.2022.12.019
Cantara A, Luo Y, Dobrovolná M, Bohalova N, Fojta M, Verga D, Guittat L, Cucchiarini A, Savrimoutou S, Häberli C, Guillon J, Keiser J, Brázda V, Mergny JL. G-quadruplexes in Helminth parasites. Nucleic Acids Res. (2022) 50, 2719-2735.https://doi.org/10.1093/nar/gkac129 Commented in https://www.polytechnique.edu/en/news/targeting-unusual-dna-structures-against-parasitic-diseases
Luo Y, Verga D, Mergny JL. Iso-FRET: An isothermal competition assay to analyze quadruplex formation in vitro. Nucleic Acids Res. (2022) 50, e93 https://doi.org/10.1093/nar/gkac465
Amrane S, Jaubert C, Bedrat A Rundstadler T, Recordon-Pinson P, Aknin C, Guédin A, De Rache A, Bartolucci L, Diene I, Lemoine F, Gascuel O, Pratviel G, Mergny JL, Andreola ML. Deciphering RNA G-quadruplex function during the early steps of HIV-1 infection. Nucleic Acids Res. (2022) 50, 12328-12343 https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkac1030/6858854
Abiri A, Lavigne M, Rezaei M, Nikzad S, Zare P, Mergny JL*, Rahimi HR. Unlocking G-quadruplexes are antiviral targets. Pharmacol Rev. (2021) 73, 897-923. https://doi.org/10.1124/pharmrev.120.000230
Cheng M, Chen J, Ju H, Zhou J, Mergny JL, Drivers of i-DNA formation in a variety of environments revealed by four-dimensional UV melting and annealing. J Am Chem Soc. (2021) 143, 7792-7807. https://doi.org/10.1021/jacs.1c02209
Cheng M, Qiu D, Tamon L, Ištvánková E, Víšková P, Amrane S, Guédin A, Chen J, Lacroix L, Ju H, Trantírek L, Sahakyan AB, Zhou J, Mergny JL. Thermal and pH stabilities of i-DNA: confronting in vitro experiments with models and in-cell NMR data. Angew Chem Int Ed Engl. (2021) 60, 10286-10294. doi: 10.1002/anie.202016801.
Miron CE, van Staalduinen L, Rangaswamy AM, Chen M, Liang Y, Jia Z, Mergny JL*, Petitjean A. Going Platinum to the Tune of a Remarkable Guanine Quadruplex Binder: Solution- and Solid-State Investigations. Angew Chem Int Ed Engl. (2021) 60, 2500-2507.



