DNA repair in prokaryotes
People involved:
- Hannu MYLLYKALLIO, DR, CNRS
- Stephane SKOULOUBRIS, MdC, Univ. Paris–Saclay
- Rima Zein-Eddine, Post-doc 'Marie Sklodowska-Curie' fellow '
- Amandine Ignace, AI, CNRS
- Wenlu Yin, Doctorant
Topic:
DNA repair systems help maintain the integrity of genomes and therefore play a key role at both the cellular level and in the transmission of a faithful genetic inheritance to offspring.
We are particularly interested in a protein called NucS (also known as EndoMS in the literature). The presence of this protein, discovered in the laboratory, is limited to the prokaryotic world. It is found in some species of Archaea (Crenarcheota and Euryarcheota) and in many bacteria belonging to the large phylum Actinobacteria (e.g. Corynebacteria and Mycobacteria). It exhibits cleavage activity against a variety of DNA substrates and is primarily involved in the detection and repair of mismatches.
Work is currently being undertaken within our team to better understand the mechanism by which NucS is involved in this repair. Two bacterial models are being studied in the lab: Corynebacterium glutamicum and Mycobacterium smegmatis. Various interdisciplinary approaches including molecular biology, genetics, biochemistry, microscopy and bioinformatics are used.
It has already been shown that NucS interacts with the protein known as beta-clamp (i.e. PCNA in archaea and DnaN in bacteria). Part of the project is therefore aimed at identifying and characterizing other partner proteins involved in this repair pathway. A second part of our work is focused on assessing the impact of mutations in the nucS gene on antibiotic resistance, which has been detected in numerous clinical isolates of the tuberculosis etiologic agent, Mycobacterium tuberculosis.
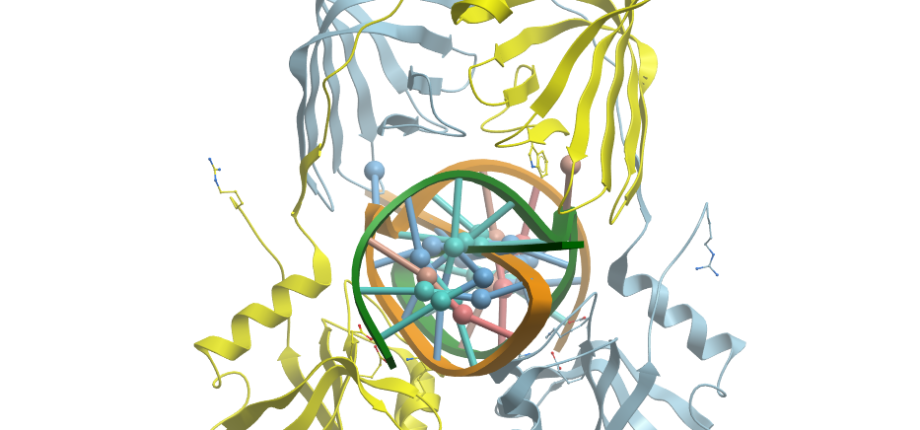
Figure. A) Dimeric 3D structure of the NucS protein from Thermococcus kodakarensis (PDB: 5GKE; 5GKJ). The protein in the absence of DNA has an "open" conformation (left) that closes in the presence of a mismatched substrate (right) (adapted from Nakae et al. (2016). Structure, 24:1960-71).
-----------------------------------------------
Keywords: microbiology, DNA repair, mismatches, NucS, beta-clamp, C. glutamicum, M. smegmatis, M. tuberculosis, antibiotic resistance.
-----------------------------------------------
Publications
Zein-Eddine R, Le Meur A, Skouloubris S, Jelsbak L, Refrégier G, Myllykallio H. NPJ Antimicrob Resist. 2025 Apr 29;3(1):35. https://doi.org/10.1038/s44259-025-00107-1
Ishino S, Skouloubris S, Kudo H, l'Hermitte-Stead C, Es-Sadik A, Lambry JC, Ishino Y, Myllykallio H. Nucleic Acids Res. 2018 Jul 6;46(12):6206-6217. https://doi.org/10.1093/nar/gky460
Rezgui R, Lestini R, Kühn J, Fave X, McLeod L, Myllykallio H, Alexandrou A, Bouzigues C. PLoS One. 2014. Nov 20;9(11):e113493. https://doi.org/10.1371/journal.pone.0113493
Creze C, Ligabue A, Laurent S, Lestini R, Laptenok SP, Khun J, Vos MH, Czjzek M, Myllykallio H, Flament D. J Biol Chem. 2012 May 4;287(19):15648-60. https://doi.org/10.1074/jbc.M112.346361
Creze C, Lestini R, Kühn J, Ligabue A, Becker HF, Czjzek M, Flament D, Myllykallio H. Structure and function of a novel endonuclease acting on branched DNA substrates. Biochem Soc Trans. 2011 Jan;39(1):145-9. https://doi.org/10.1042/BST0390145
Ren B, Kühn J, Meslet-Cladiere L, Briffotaux J, Norais C, Lavigne R, Flament D, Ladenstein R, Myllykallio H. Structure and function of a novel endonuclease acting on branched DNA substrates. EMBO J. 2009 Aug 19;28(16):2479-89. https://doi.org/10.1038/emboj.2009.192
Ren B, Kuhn J, Meslet-Cladiere L, Myllykallio H, Ladenstein R. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007 May 1;63(Pt 5):406-8. https://doi.org/10.1107/S1744309107015278
Meslet-Cladiére L, Norais C, Kuhn J, Briffotaux J, Sloostra JW, Ferrari E, Hübscher U, Flament D, Myllykallio H. J Mol Biol. 2007. Oct 5;372(5):1137-48. https://doi.org/10.1016/j.jmb.2007.06.056
Collaborations
- Dr Guislaine Refrégier, IDEEV, UniV. Paris-Saclay
- Pr Emmanuelle Cambau, University Paris Cite - APHP (Bichat Claude-Bernard hospital)
- Pr Lars Jelsbak, DTU, Danemark
- Pr Pierre Leblond, Univ. de Lorraine, Nancy
- Didier Flament, IFREMER, Brest
Fundings
- ANR-22-CE12-0042 / Avril 2023, durée 48 mois
- ANRS – Maladies infectieuses émergentes Agence autonome de l’INSERM (EMERGENCES N°23534)



