Replication in archaea

Staff
Roxane Lestini / Ecole polytechnique
Nicolas Olivier / CNRS
Stéphane Duigou / CNRS
Lionel Guittat / Université Sorbonne Paris-Nord
Caroline l'Hermitte-Stead / INSERM
Tom MARIOTTE / Post-doc
Kate Sorg / PhD
Internship proposals / Propositions de stage :
Please do not hesitate to contact us if you are interested in joining us.
Topic
Little is known about the regulation of replication of polyploid archaeal genomes. As for polyploid bacteria, it is very likely that not all copies of the chromosome are replicated. Remarkably, in most species of polyploid archaea the circular chromosome contains multiple replication origins, a feature only observed for the linear chromosomes of eukaryotes. These archaea provide a unique cellular context in which it is very interesting to understand the dynamics of replication.
We are using the salt-loving archaea Haloferax volcanii. Because of the significant heterogeneity in the genome copy number per cell within a population, it is crucial to include methods allowing single-cell resolution to investigate replication dynamics.
We are investigating replication dynamics in H. volcanii cells by studying the localization of the replication protein Rpa2. Rpa2 localization in cells expressing Rpa2 fused to the GFP was monitored in live cells by conventional and super-resolved microscopy 3D-SIM techniques (Delpech et al., 2018) and revealed replication foci (RF) in living archaea cells for the first time.
We are now developing strategies to study replication foci using super-resolution microscopy techniques on both fixed and live cells to (i) map as accurately as possible all replisomes per cell, (ii) correlate the number of replisomes and the ploidy level per cell, and (iii) characterise how replication regulation is influenced by the environment and metabolism. This includes:
- targeting Rpa2 proteins in fixed cells by super-resolution STORM microscopy (STochastic Optical Reconstruction Microscopy)
- targeting Rpa2 proteins in live cells using a combination of conventional and super-resolved fluorescence microscopy (SIM, PALM)
- Nascent DNA labelling through BrdU incorporation followed by specific immunodetection using superresolution STORM technique (Lestini, 2022)
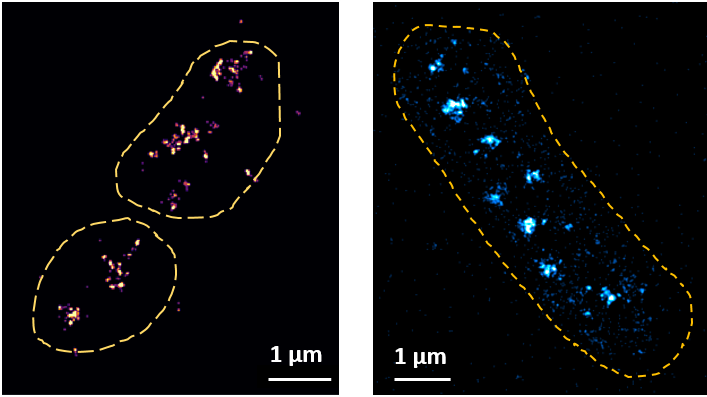
Figure: (left panel) STORM image of FLAG::Rpa2 signal, (right panel) Live-PALM image of Dendra2::Rpa2
Selected recent publications
Lestini, R., Collien, Y., Olivier, D., Olivier, N., Myllykallio, H. (2022). BrdU Incorporation and Labeling of Nascent DNA to Investigate Archaeal Replication Using Super-Resolution Imaging. Methods Mol Biol 2522:419-434.
Cockram, C., Thierry, A., Gorlas, A., Lestini, R., and Koszul, R. (2021). Euryarchaeal genomes are folded into SMC-dependent loops and domains, but lack transcription-mediated compartmentalization. Mol Cell 81, 459-472 e410.
Delpech F., Collien Y., Mahou P., Beaurepaire E., Myllykallio M., Lestini R. (2018) Snapshots of archaeal DNA replication and repair in living cells using super-resolution imaging, Nucleic Acids Research, 46(20), 10757–10770
Lestini, R., Laptenok, S.P., Kuhn, J., Hink, M.A., Schanne-Klein, M.C., Liebl, U. and Myllykallio, H. (2013) Intracellular dynamics of archaeal FANCM homologue Hef in response to halted DNA replication. Nucleic Acids Research, 41, 10358-10370.
Lestini, R., Duan, Z. and Allers, T. (2010) The archaeal Xpf/Mus81/FANCM homolog Hef and the Holliday junction resolvase Hjc define alternative pathways that are essential for cell viability in Haloferax volcanii. DNA Repair (Amst), 9, 994-1002.
Lestini, R. and Michel, B. (2007) UvrD controls the access of recombination proteins to blocked replication forks. EMBO J, 26, 3804-3814.
Main ongoing collaborations
- Romain Koszul, Institut Pasteur
- Sébastien Ferreira-Cerca, Ecole polytechnique
- Amy Schmid, Duke University (US)
- Jörg Soppa, Goethe University Frankfurt (Germany)
- Thorsten Allers, Nottingham University (UK)
Alumni/PhD students
Titouan Bouvier d'Yvoire / M2 student
Tom Garnier / Master 2 student
Zoé Commander / Master 1 student
Yoann Collien / PhD student (2016-2019)
Floriane Delpch / PhD student (2013-2016)



